2022.07.02.16
Files > Volume 7 > Vol 7 No 2 2022
Isolation and molecular identification of Exiguobacterium profundum from drain water and study of some physiological properties
1,2,3 Department of Biology, College of Science, University of Mosul, Iraq
Corresponding Author: [email protected]
Available from: http://dx.doi.org/10.21931/RB/2022.07.02.16
ABSTRACT
Some microorganisms' stress tolerance and adaptability through the ability to live in new environments previously considered hostile to dense microbial growth. The aim of the study: In this work, the isolation, molecular identification, and analysis of some physiological properties of Exiguobacterium profundum isolated from washing machine drain water were carried out. Fifteen samples were collected from different washing machines' drain water, and 42 isolates were characterized and identified. Based on the morphological assay and 16S rRNA gene sequencing, one isolate was designated as Exiguobacterium profundum Kh-Am2 and was deposited in the Gene bank database under the accession number MW447498.1. The physiological properties assay showed that E. profundum could not decompose red blood cells while proving their ability to decompose crude and coconut oil, producing lipopeptide biosurfactants. It has also demonstrated its ability to form biofilm using the tube method, which helps it to live in harsh environments such as the environment of washing machines. The results showed that E. profundum is halotolerant bacteria with its ability to grow at a concentration of ( 75) g/l NaCl and the best growth was at( 25) g/l NaCl concentration. The results also showed the effect of different concentrations of NaCl on E. profundum, where the best movement rate was at a concentration of ( 25 )g/l NaCl, while the movement was completely inhibited at a concentration of (75)g/l NaCl. We have concluded that E. profundum possesses traits that enable it to live in harsh environments.
Keywords. Exiguobacterium profundum, drain water, biosurfactant
INTRODUCTION
Exiguobacterium is a bacterial genus with a wide range of species obtained from various habitats. Exiguobacterium first was recognized as a genus about three decades ago, and there are presently Seventeen species in this genus. Exiguobacterium is a halotolerant bacteria that has piqued the interest of many researchers as a crucial agent in salinity stress reduction, capable of thriving in a wide range of temperatures and pH 1.
Exiguobacterium isolates have been employed in biotechnology and industrial applications such as enzyme synthesis, bioremediation, and the breakdown of toxic materials released into the environment. Other isolates can promote plant development, and they are currently being investigated to enhance agricultural production. In the genomic sequences of diverse strains of this species, many genes encoding products crucial to agriculture and the environment have been found. In the genomic sequences of various strains of this species, many genes encoding products important to agriculture and the environment have been found. Many strains also feature stress-response genes, allowing them to colonize and thrive in a wide range of ecological settings1,2 . Exiguobacterium species such as "E. sibiricum , E. alakliphilum, , E. undae, E. oxidotolerans, E. mexicanum , E. antarkticum, E. profundum, and E. indicum" have been discovered in a variety of environments3 .
E. profundum is a species of Exiguobacterium, Gram-positive, nonsporulating rods that can be found mono, pairs or in short motile chains with peritrichous flagella. Colonies are spherical (1–2 mm) and creamy or orange in color under anaerobic or aerobic conditions. Chemo-organ trophic and facultatively anaerobic. Positive for catalase and negative for oxidase. It is slightly thermophilic (growing within 12 - 49 °C, optimum at 45 °C) and halotolerant (growth in 11 percent NaCl, optimal 0–2 percent NaCl). Its pH range for growth is 5.5–9.5. (Optimum pH 7.0). Dextrose, lactose, maltose, mannose, and sucrose were used to test carbohydrate fermentation. The major organic acid produced by carbohydrate fermentation is L-Lactate, with traces of formate, ethanol, and acetate also present. Mueller Hinton broth, "Soybean Casein Digest broth, King's broth, Mac Conkey broth, and Nutrient broth media" were used to assess the maximum growth and pigment production properties 4,5.
According to several studies, Exiguobacterium profundum can be used in bioremediation and toxic degradation by producing lipopeptide biosurfactants, which have stable chemical characteristics, are renewable, environmentally beneficial, and have low toxicity 6,2,7.
Currently used in industry, cosmetics, and pharmaceutical emulsions, foaming agents, wetting agents, emollients, and have antibacterial activity against gram-negative bacteria 8,2,9.
This research aimed to isolate and molecular identify Exiguobacterium profundum and study some physiological properties such as biosurfactant production, biofilm production, and salinity on growth and motility.
MATERIALS AND METHODS
Sample Collection and Isolation
Fifteen samples were collected from washing machines, drained water in sterile containers and brought to the laboratory. Following, 0.1 mL from each sample was cultured on Trypto Soy Agar (TSA) by using sterile swab sticks (BIONOVO, Poland) . The plates were incubated at 37°C for 24-48 h. After the incubation, isolates were purified to obtain single colonies, then underwent to identification by using Gram-stain, colonies appearance and molecularly identified by using the 16S rRNA gene and the following assays 10.
Amplification of 16S rRNA gene and Phylogenetic Tree
DNA extraction
The isolate was grown at 37 °C for 24 h on a rotary shaker (250 rpm) in a falcon tube containing 20 mL of Tryptic Soy Broth (TSB). The method for extracting DNA was subsequently followed using the G- spin DNA extraction kit (intron biotechnology, Korea). The 16S rRNA genes were amplified by polymerase chain reaction (PCR) from genomic DNA of selected isolates according to the Maxime PCR (PreMix kit) methodology. For PCR reaction, 1.5µl of DNA template, 1μl of 10 picomols/µl of each conserved primers 1250R (5'- GGTTACCTTGTTACGACTT- 3') and 1250 F (5'- AGAGTTTGATCCTGGCTCAG- 3') from (Integrated DNA Technologies company, Canada), 5µl of Taq PCR PreMix and 16.5 µl ddH2O for a total volume of 25μl. PCR with the optimal cycle setting for the detecting 16S rRNA gene was carried out using a Bio-Rad MyCycler thermal cycler (Bio-Rad, USA) with an initial denaturation step at 95oC for 5 min, Denaturation step at 95oC for 45 sec, 35 cycles of 95oC for 45 sec, 58oC for 45 min, 72oC for 45 sec, followed by a final extension step of 72oC for 7 min. Amplification products were analyzed by electrophoresis in 1% (w/v) agarose gel.
Purification of PCR Products
According to the manufacturer's instructions, the PCR products were purified using the QIAquik PCR purification kit (QIAGEN). In brief, one volume of PCR product mixed with five volumes of Binding Buffer was mixed thoroughly and, after that, placed in a QIAquick spin column, followed by spinning for 60 seconds to bind the DNA, flow-through was removed, the column was then placed back into the collecting tube. DNA was centrifuged for 60 seconds after being cleaned with 0.75 ml of washing buffer. The flow was removed, and the tubes were centrifuged for another minute. After that, the column was put in a fresh 1.5 ml centrifuge tube. Fifty µl Buffer of Elution was added to the middle of the QIAquik membrane for DNA elution, and the tubes were centrifuged for 1 minute. Macro gene/Korea Sequencing Service sequenced the purified PCR products. (Following Kit instructions)
Phylogenetic tree
The DNA sequence was collected and matched to sequences in the gene bank databases. Homolog surveys were carried out using internet bioinformatics tools (www.ncbi.nlm.nih.gov.BLAST). Sequences had a lot of similarities (100%) with species E. profundum. In FASTA format, all isolates were retrieved and downloaded. A maximum-likelihood Multiple Sequence Comparative by Log Expectation (MUSCLE) software created phylogenetic trees with the 16S gene of Kh-Am2 strain using program MEGA version 7.0 and matched sequences. The confidence value of individual branches was calculated using 1000 bootstrap copies of the original sequences. The complete 16S rRNA gene sequences were deposited in the Gene Bank database.
Biosurfactant screening
Blood agar hemolysis assay
Sterile fresh blood in volume 1.5 mL was added to a 100 mL Erlenmeyer flask containing a sterile blood base medium. The selected isolate was streaked onto blood agar and cultured for 48-72 hours at 37°C 9.
Oil spreading assay
The method of 9 was used in the screening of biosurfactants production for Kh-Am2 isolate. Using a petri dish, 30 mL of distilled water was poured into it, 1 ml of coconut oil and crude oil were separately tubed and dropped into the center; in the center of the oil layer, 20 mL of the Kh-Am2 culture was placed.
Spread Plate assay
Following the method of Spread Plate assay with modified10, the assay was done to conduct the ability of the selected isolate to consume crude oil and coconut oil. The plates containing Bushnell Hass medium (BHM) as a mineral salt medium comprised of (K2HPO4 1gm, MgSo4.7H2O 0.2 gm, FeCl3 0.05 gm, CaCl2 0.02 gm, NH4NO3 1gm, , Agar 15 gm) in 1L of distal water were prepared with added 1% of crude oil and coconut oil separately. Each plate was inoculated by 0.1 ml 1.5×108 cells ml−1of selected isolate suspension. Then plates were incubated for seven days at 37C˚. The growth of colonies after the incubation time is taken as a sign of their ability to use crude and coconut oil as a source of energy and carbon.
Biofilm formation detection using the tube method
The chosen isolate was inoculated in a tube containing ten trypticase soy broth and 1% glucose and cultured for 24 hours at 37°C. After incubation, tubes were poured and washed with phosphate buffer saline (pH 7.3) before drying. The tube was then colored with crystal violet (0.1 percent ). Any residual discoloration was removed with deionized water and then dried inside out. The findings of the control were used to evaluate the tube method. It was considered a positive sign of biofilm development when a visible film lined the tube's wall and bottom. 11,12.
Salinity tolerance
The Kh-Am 2 isolate was cultivated on YP culture media with NaCl concentrations of (0), ( 25 ) , (50 ) and (75) g/l. 75 hours at 25°C incubation with orbital agitation at 120 rpm. OD600 was measured in a UV–visible spectrophotometer UV 1800 to track growth.
Salinity's Influence on Motility
The selected isolate was cultivated on YP agar (0.3% agar / Difco) . A drop over the agar in the Petri plate's middle inoculated. The motility ratio were evaluated after 72 hours of incubation at 25 °C in ( 0 ) , (25) , (50 )and (75 ) g /l NaCl13 .
RESULTS AND DISCUSSION
The streaking results on cultured medium plates revealed that the drain water samples contain various microbial communities with different gram-positive and gram-negative bacteria. A total of 42 bacterial isolates were isolated. The target Gram-positive rods bacteria of Exiguobacterium was the most numerous of other isolates, and this agrees with what was indicated by both4,14 and was chosen for further studies
16S rRNA gene sequence and phylogenetic analysis
The isolate designated as the Kh-Am2 and their 16SrRNA gene sequences was recorded in the NCBI Gene bank databases under the accession number MW447498.1. Once the 16S rRNA gene of the Kh-Am2 isolate was amplified and analyzed using a 1 percent agarose gel-electrophoretic method, the PCR result was 1500 bp, as shown in Gene data (http://www.ncbi.nlm.nih.gov) a search tool was used to perform BLAST analysis on partial 16S r RNA sequences. The Kh-Am2 isolate exhibited 99-to-100% matched with the NCBI Gen database's sequences. The isolate's Neighbour Joining tree as constructed in MEGA version 7
(Figure 1), identified that a neighbor-joining tree based on similar 16S rRNA gene sequences for the E. profundum Kh-Am2 is the most closely related to Exiguobacterium sp. 8A. They are both clustered with E. profundum strain fsznc-03 and E. profundum strain GN31a.
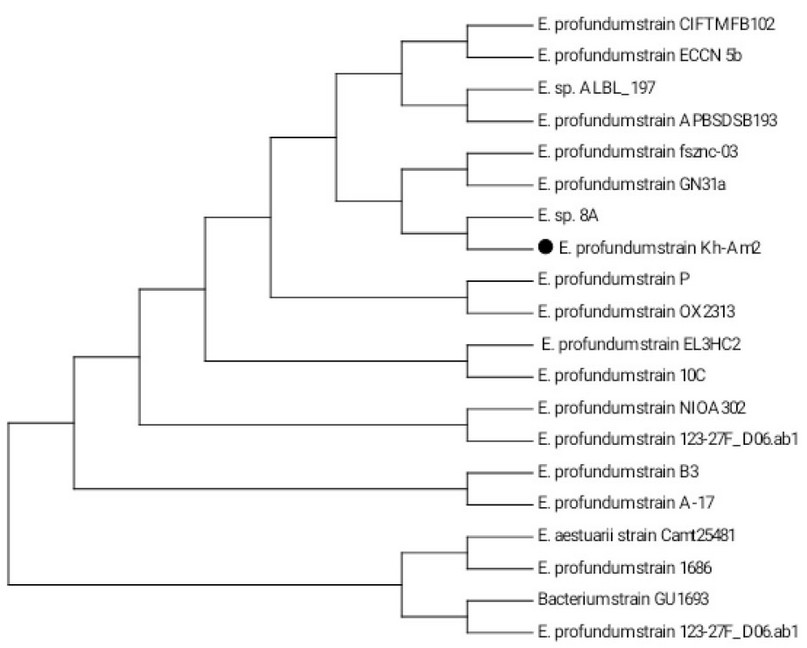
Figure 1. Phylogenetic tree (1000 bootstrap) constructed by MEGA7 tool from 19 merged nucleotide sequences of 16S rRNA gene homologous to that of Exiguobacterium profundum strain Kh-Am2 (indicated by black cycle).
Blood agar hemolysis assay
The results of the hemolysis test on the blood agar medium showed a negative result indicating that no clear areas were formed around the inoculation line, which is consistent with what was indicated by both 9 as it does not produce hemolysin that breaks down the red blood cells.
Oil spreading assay
The results showed the ability of Kh-Am2 isolate to produce biosurfactant in terms of clear zones and oil displacement zones for both types of crude oil and coconut oil, as shown in (figure 2). The reason is due to the ability of Kh-Am2 to produce biosurfactant; in addition, Exiguobacterium strains produce alkali esterase enzyme
belonging to Lipolytic enzymes 22, and this is consistent with what was referred to by 9, who stated that The biosurfactant produced by E. profundum could be classified into the lipopeptide group, It also gave the results of the oil spreading assay presented by Ahmed's observation for Bacillus cereus ND 1, as it showed its ability to decompose crude oil 15, Biosurfactants work as emulsifiers by lowering surface tension whereas, biosurfactants are made up of polar and nonpolar parts16 .
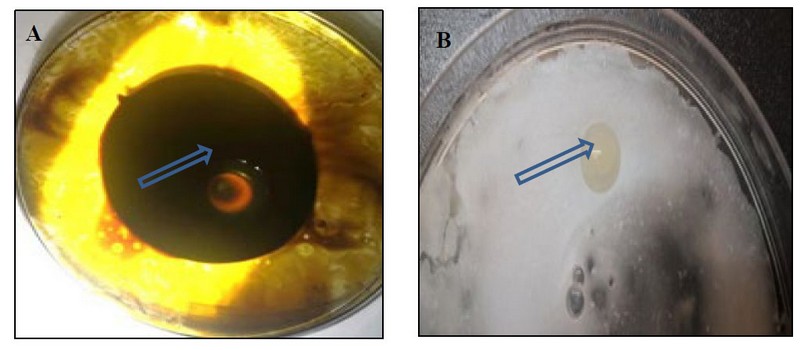
Figure 2. The clear zone begins to form in the oil spreading assay:- (A) crude oil (B) coconut oil
Spread Plate assay
Kh-Am2 isolate showed its ability for growing on BHM containing crude oil and coconut oil separately, which indicates its ability to consume both crude oil and coconut oil, as shown in (figure 3), due to its ability to use crude oil and coconut oil as a source of energy and carbon. The researcher17 pointed out the role of Exiguobacterium sp. AO-11 in removing crude oil pollutants from the microcosms of sediments, and The researcher Chen referred to the role of Exiguobacterium sp in the decomposition of crude oil.18
This could be a unique trait that helps to the survival of these populations and reflects the ability of these creatures to thrive in such harsh conditions in the washing machines, as it can depend on its growth on oils sticking to clothes when it comes out with drain water.
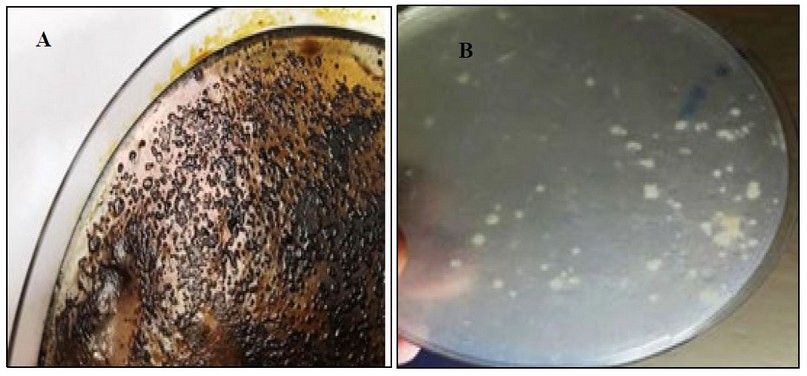
Figure 3:- The ability of Kh-Am2 isolate to consume crude and coconut oil on BHM :- A(BHM with 1% crude oil) B (BHM with 1% coconut oil)
Biofilm formation detection using the tube method
The results showed the ability of Kh-Am2 to adhere and form a biofilm by using the tube method in terms of the deposition of the crystal violet on the inner side of the test tube, as shown in (figure 4). Biofilms are a community of bacteria that produce extracellular substances such as proteins, polysaccharides, DNA, RNA, and water. The essential component of a biofilm is the nutrient flow control matrix19. The ability of Kh-Am2 isolate to generate biofilms was demonstrated using the biofilm tube approach. Where the researcher17 was referring to Exiguobacterium sp. ability to create biofilm. Biofilm production is one of the most important mechanisms that the Kh-Am2 isolate has for surviving in harsh settings.

A:- Positive result B:-Control
Figure 4:- Biofilm formation by Kh-Am2 on the inner sides of the test tube
Salinity tolerance
The results showed the ability of Kh-Am2 isolate to grow in high concentrations of salt. The best growth was found in (25) g/l of NaCl, where the absorbance reached 0.8 at OD600 nm. The concentrations (0), (50), (75) g/l of NaCl were 0.4, 0.3, 0.3 at OD600, respectively, as shown in (figure 5), where it gave a similar reading for both concentrations (50) and (75) g/l of NaCl, this result makes the Kh-Am2 isolate is classed as halotolerant bacteria, While the researcher13 indicated that Exiguobacterium sp. SH31 were unable to grow at a concentration of (75) g/l NaCl percent, but they were able to grow at a salt concentration of( 50) g /l NaCl percent 19 also noted that the growth of E. profundum PHM11 stopped at the concentration of 2500 mM. There is little published research on how Exiguobacterium tolerates salinity by fine-tuning its gene expression patterns and modifying its inherent physiology. Living in extreme environments causes their genes to adapt to harsh environmental conditions, and thus the enzymes and proteins encoded by the genes can withstand extreme conditions.
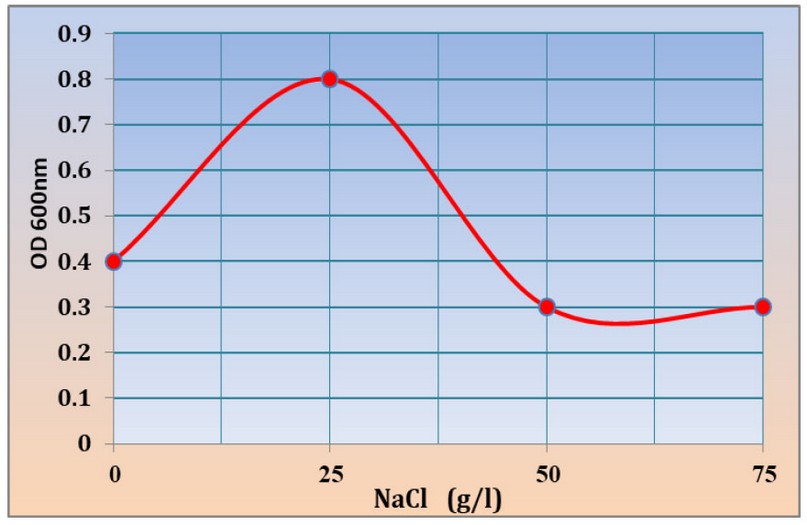
Figure 5:- Growth of Kh-Am2 in 0, 25, 50 and 75 g/l NaCl
Salinity's Influence on Motility
The results showed that Kh-Am2 isolate moved (swimming movement) on YP agar at (0) , (25) and (50) g/l NaCl, where the highest percentage of movement was observed at a salt concentration of (25) g/l NaCl, which represented the best growth for the isolate as observed in (Figure 6). At the same time, the motility was wholly inhibited at ( 75 ) g/l NaCl as shown in (figure 6).
Some bacteria respond to osmotic pressure, as in Pseudomonas putida21. Although Exiguobacterium bacteria are described as motile22, it suggests that specific salt concentrations make the cell less active or could be governed by environmental factors somehow.23 Flagella synthesis requires much energy, and bacteria can block specific processes in reaction to stress. Motility was reduced, and biofilm development was inhibited.13The results of salinity on motility showed that the highest motility (Swimming motility) was obtained at a concentration of (25) g/l NaCl. In contrast, motility was inhibited at a concentration of( 75) g/l NaCl, and this agrees with the scientist (13) in obtaining the highest percentage of motility at a concentration of( 25) g/l NaCl for Exiguobacterium sp. SH31 while the movement was inhibited at a (50) g/l NaCl concentration.
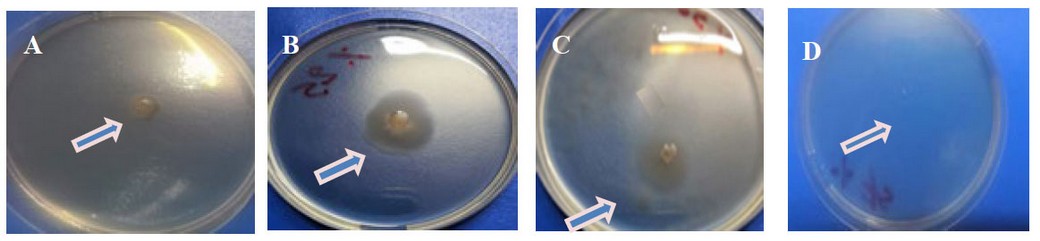
Figure 6:- Effect of salinity in the motility of Kh-Am2 with different concentrations of NaCl after incubation 72 h at 25°C. A .Concentration 0 g/l of NaCl, B.Concentration 25 g/l of NaCl,
C .Concentration 50 g/l of NaCl, D.Concentration 75 g/l of NaCl.
CONCLUSION
Under extreme conditions, salinity is a significant element in determining the spread of life. Our study suggests that E. profundum Kh-Am2 possesses a stressing regulation system that controls gene expression and supports the organism's survival and growth in salt environments. Our data revealed the potential of the E. profundum Kh-Am2 isolate to grow at a high salt concentration of 75 g/L and the ability to develop a biofilm. This was related to maintaining the integrity of bio-molecules in the physiological process and preserving cell architecture and function. E. profundum Kh-Am2 can degrade crude and coconut oil in an oil spreading assay, indicating that the bacteria can be used in bioremediation and toxic degradation.
More analyses are needed to comprehend further the gene regulation of salinity pathway of the E. profundum Kh-Am2 isolated from waste water of a washing machine.
Acknowledgment:
The researchers would like to express their gratitude to the University of Mosul for supporting this study. The tools were also provided by the Biology department and the Iraqi Ministry of Higher Education and Scientific Research.
Conflict of interest
The authors declare that they have no conflict of interest.
Funding
This research did not receive any specific support from public or commercial funding agencies.
REFERENCE
1. Rodrigues, D. F., Ivanova, N., He, Z., Huebner, M., Zhou, J., & Tiedje, J. M. Architecture of thermal adaptation in an Exiguobacterium sibiricum strain isolated from 3 million-year-old permafrost: a genome and transcriptome approach. BMC genomics,2008؛ 9(1), 1-17.
2. Kasana, R. C., & Pandey, C. B. Exiguobacterium: an overview of a versatile genus with potential in industry and agriculture. Critical reviews in biotechnology,2018؛ 38(1), 141-156.
3. Bharti, N., Yadav, D., Barnawal, D., Maji, D., & Kalra, A. Exiguobacterium oxidotolerans, a halotolerant plant growth promoting rhizobacteria, improves yield and content of secondary metabolites in Bacopa monnieri (L.) Pennell under primary and secondary salt stress. World Journal of Microbiology and Biotechnology,2013؛ 29(2), 379-387.
4. Crapart, S., Fardeau, M. L., Cayol, J. L., Thomas, P., Sery, C., Ollivier, B., & Combet-Blanc, Y. Exiguobacterium profundum sp. nov., a moderately thermophilic, lactic acid-producing bacterium isolated from a deep-sea hydrothermal vent. International journal of systematic and evolutionary microbiology,2007؛ 57(2), 287-292
5. Manon, M. V., Keerthana, G., & Preethi, K. Evaluation of antioxidant potential of bioactive colored metabolite isolated from Exiguobacterium Profundum BC2-11 and it's bioactivities. Int J Recent Sci Res,2015؛ 6(4), 3612-3617
6. Ciccyliona, D. Y., & Nawfa, R. Pengaruh pH terhadap produksi biosurfaktan oleh bakteri Pseudomonas aeruginosa lokal. Jurusan Kimia, FMIPA-ITS, Surabaya, 2012؛1-6
7. Kubicki, S., Bollinger, A., Katzke, N., Jaeger, K. E., Loeschcke, A., & Thies, S. Marine biosurfactants: biosynthesis, structural diversity and biotechnological applications. Marine drugs,2019؛ 17(7), 408
8. Md, F. Biosurfactant: production and application. J Pet Environ Biotechnol,2012؛ 3(4), 124
9. Setiani, N. A., Octaviyani, W., Hamdani, S., & Mardiah, I.Studies On Biosurfactant Produced Using Exiguobacterium Profundum. Acta Biochimica Indonesiana,2020؛2(2), 325922.
10. Younus, R. M., Aziz, E. M. T., & Mohammed, D. A. Degradation of hydrocarbon substances by some bacterial species isolated from contaminated soils with motor oil. EurAsian Journal of BioSciences,2020؛ 14(1), 1087-1095.
11. Hassan, A., Usman, J., Kaleem, F., Omair, M., Khalid, A., & Iqbal, M. Evaluation of different detection methods of biofilm formation in the clinical isolates. Brazilian journal of infectious Diseases,2011؛ 15(4), 305-311.
12. Sultan, A. M., & Nabiel, Y. Tube method and Congo red agar versus tissue culture plate method for detection of biofilm production by uropathogens isolated from midstream urine: which one could be better?. African Journal of Clinical and Experimental Microbiology,2019؛ 20(1), 60-66.
13. Remonsellez, F., Castro-Severyn, J., Pardo-Esté, C., Aguilar, P., Fortt, J., Salinas, C., ... & Saavedra, C. P. Characterization and salt response in recurrent halotolerant Exiguobacterium sp. SH31 isolated from sediments of Salar de Huasco, Chilean Altiplano. Frontiers in microbiology,2018؛ 9, 2228.
14. Hu, N., Zou, W., Cai, Q., Liu, Y., Chen, K., Li, M., ... & Zeng, L. The First Report of Cerebral Nocardiosis Caused by Nocardia terpenica Together With Exiguobacterium profundum Bacteremia. Jundishapur Journal of Microbiology,2018؛ 11(10).
15. Daxini, N., & Mistry, K. Biosurfactant assistance in crude oil degradation by halophilic Bacillus cereus ND1,2018.
16. Sanches, M. A., Luzeiro, I. G., Alves Cortez, A. C., Simplício de Souza, É., Albuquerque, P. M., Chopra, H. K., & Braga de Souza, J. V. Production of Biosurfactants by Ascomycetes. International Journal of Microbiology, 2021.
17. Chauhan, D., Agrawal, G., Deshmukh, S., Roy, S. S., & Priyadarshini, R. Biofilm formation by Exiguobacterium sp. DR11 and DR14 alter polystyrene surface properties and initiate biodegradation. RSC advances,2018؛8(66), 37590-37599.
18. Chen, Q., Li, J., Liu, M., Sun, H., & Bao, M. (2017). Study on the biodegradation of crude oil by free and immobilized bacterial consortium in marine environment. PloS one, 12(3), e0174445.
19. Muhsin, J., Ufaq, T., Tahir, H., & Saadia, A. Bacterial biofilm: its composition, formation and role in human infections. Journal of Microbiology and Biotechnology,2015؛ 4, 1-14.
20. Patel, V. K., Srivastava, R., Sharma, A., Srivastava, A. K., Singh, S., Srivastava, A. K., ... & Saxena, A. K. Halotolerant Exiguobacterium profundum PHM11 tolerate salinity by accumulating L-Proline and fine-tuning gene expression profiles of related metabolic pathways. Frontiers in microbiology,2018؛ 9, 423.
21. Bojanovič, K., D'Arrigo, I., & Long, K. S. Global transcriptional responses to osmotic, oxidative, and imipenem stress conditions in Pseudomonas putida. Applied and environmental microbiology,2017؛83(7), e03236-16.
22. Amaresan, N., Kumar, M. S., Annapurna, K., Kumar, K., & Sankaranarayanan, A. (Eds.). Beneficial Microbes in Agro-Ecology: Bacteria and Fungi. Academic Press,2020.
23. Chatterjee, A., Cui, Y., Chakrabarty, P., & Chatterjee, A. K. Regulation of motility in Erwinia carotovora subsp. carotovora: quorum-sensing signal controls FlhDC, the global regulator of flagellar and exoprotein genes, by modulating the production of RsmA, an RNA-binding protein. Molecular plant-microbe interactions,2010؛ 23(10), 1316-1323.
Received: 30 November 2021 / Accepted: 25 January 2022 / Published:15 May 2022
Citation: G.O. Al-Ani A, Mohammed Younis K, M.Y. Al-Taee S. Isolation and molecular identification of Exiguobacterium profundum from drain water and study of some physiological properties. Revis Bionatura 2022;7(2) 16. http://dx.doi.org/10.21931/RB/2022.07.02.16
