2022.07.04.58
Files > Volume 7 > Vol 7 No 4 2022
1. Department of Cellular and Molecular Biology, Faculty of Biological Sciences, Islamic Azad University- Tehran North Branch, Tehran, Iran. PO Box: 19585/466, Telephone: +98212565149, Fax: +982177009847;
2. Reproductive Immunology Department, Avicenna Research Institute, ACECR, Shahid Beheshti University of medical sciences, Tehran, Iran. Telephone: 02122432020, Fax: +98(21)22432021
* Corresponding author: Email: [email protected]
Available from: http://dx.doi.org/10.21931/RB/2022.07.04.58
ABSTRACT
Exosomes naturally carry the biomolecules in the body; they perform this task efficiently without compromising the immune system and by breaking through all the biological barriers, so they can be the best choice for designing and introducing a drug and gene transfer system. Extraction of the exosomes from the cell culture medium was performed by precipitation with an Exoquick kit solution. Nanoparticle specificity analysis was performed using scanning electron microscopy and dynamic light scattering. Trizol reagent (Invitrogen) was used for RNA extraction. Single-strand cDNA synthesis was performed from the miRNA and RT-PCR. Data were analyzed using a threshold cycle comparative method and cell cycle analysis using flow cytometry. Exosomes containing miR-144 can dramatically decrease the expression level of crucial TGF-β pathway genes, SMAD4 and TGF-βR2, in breast cancer cells. Botulinum toxin A inhibits cancer cell growth by inhibiting the TGF-β pathway. The simultaneous combination of engineered exosomes containing miR-144 and bacterial botulinum toxin A has increased effects on inhibiting the TGF-β signaling pathway. It causes cell cycle arrest in breast cancer cells. The present study's findings showed that overexpression of miR-144 in breast tumor cells results in the packaging of miRNA in exosomes derived from these cells. As a result, the exosomal platform for nucleic acid transfer to the cell appears to be an effective transducer for gene transfer to the cell. It could be used as a suitable adjunct to cancer therapeutic studies.
Keywords: Breast cancer, botulinum toxin A, exosome, miRNA, biomarker.
INTRODUCTION
Breast cancer is the second most common malignancy in the world. Breast cancer is the most common cause of death in women less than 45 years of age. Statistics show that cancer is rare in the age group of 20 to 24, but it is more prevalent in women aged 34 to 39. Modifiable and non-modifiable risk factors in breast cancer predictor models of body mass index, BRCA1 and BRCA2 mutations, Parity Li Fraumeni syndrome, high alcohol intake, lifestyle, radiation exposure in utero, breastfeeding, and smoking have been noted. Studies in the molecular, histopathological, genetic, and genomic domains have shown that young women with breast cancer have an increased incidence of more invasive subtypes with worse overall prognosis, increased genetic susceptibility, differential tumor genes, and genomic-specific signatures1.
The miRNAs are a group of 25 nucleotide-long RNAs. They are small non-coding RNAs that approximately 18 normally regulate gene expression at the post-transcriptional stage by binding to the 3'UTR portion of a target mRNA. Studies have shown that alterations in miRNA expression occur in a range of cancers. MiRNAs also control cancer-related processes such as proliferation, apoptosis, migration, and invasion. Recent studies have also shown that miRNAs play a key role in stem cell differentiation. The role of miRNAs as new predictive and prognostic markers has been considered in several studies and has been prominent as potential therapeutic targets2. MiR-548c-5p, miR-181d, miR-487b, miR-206, miR-195, miR-30d, miR-149, miR-183, miR-182 and miR-320, miR-10a, miR-130, miR-127-3p, miR-143, miR-10b, miR-125b, and miR-195 are used as molecular signatures early in the onset of breast cancer3.
In this study, we aimed to evaluate the regulation pattern of MiR-144 in response to exosome therapy of breast cancer cells, as well as evaluate the breast cancer cells micro-environment.
MATERIALS AND METHODS
Cell culture
The cell line of MDA-MB-231 (invasive breast cancer cell line) was bought from the Pasteur Institute, Iran. The cells were cultured in a DMEM medium containing 11% fetal calf serum (FBS), 1mM L-glutamine, 100 unit/ ml penicillin, and 100μg /ml streptomycin in wet incubation; they were grown with 5% CO2 at 37 °C.
Cell passage of adherent cells was performed by evacuating the cell culture medium, washing twice with PBS, adding 2ml of trypsin solution, and incubating at 37 °C for 3 to 5 minutes. After transferring to Falcon, the cell suspension was centrifuged at 1500rpm for 5min. The resulting precipitate was dissolved in 1 ml of a complete medium. The resulting suspension was divided between 2 to 3 new flasks. Cell culture flasks were incubated and maintained at 37 °C with 5% carbon dioxide.
Exosome Isolation from Cell Culture Using Sedimentation with ExoQuick Kit
The culture medium or cell supernatant collected at different passages was used to isolate the exosomes based on the manufacturer's guidelines4.
Scanning electron microscopy (SEM)
A small volume of the exosome was purified and washed with glutaraldehyde 2.5% and fixed with PBS. The sample was then dialyzed with ethanol and dried on a glass surface with a thin layer of gold. The size and morphology of the exosomes were assessed by scanning electron microscopy (Digital SEM, KYKY-EM3200, China).
Dynamic light scattering (DLS)
The volume of 40μL of extracted exosomes that had been dissolved in PBS reached 300μL by PBS. Then the solution was sonicated. Exosome size was measured by the Zetasizer Nano ZS (Malvern Instruments, UK) apparatus.
Overexpression of microRNA
The tube containing the lyophilized microRNA precursor sequence was briefly centrifuged to collect all material at the end of the tube. The precursor sequence was dissolved in 331μl of nuclease-free water according to the manufacturer's proposed protocol, increasing this amount to give a 11μM solution. The tube was kept at room temperature for a few minutes, and then the contents of the tube were mixed with gentle pipetting. The resulting suspension was stored at -20 °C.
Total RNA Extraction
In the present study, the Trizol (Invitrogen) reactor was used for RNA extraction. The manufacturer's guideline was followed to isolate RNA from the cell or exosome: they were first centrifuged in a Falcon tube and then added to the pellet containing 1ml of Trizol reagent and incubated at room temperature for 5min. 200µl chloroform per 1ml Trizol was added to the mixture. The micro-tube centrifugation was performed at 12000g at 4 °C for 15min. Isopropanol was added to the volume of the transferred fluid and incubated on ice for 20 minutes. The supernatant was discarded, and 1 ml of 75% ethanol was added to the precipitate. The microtube was vortexed and continued until the sediment was separated from the bottom of the microtube. The sample was centrifuged at 7500g for 8 minutes at 4 °C5.
Statistical analysis
The data were presented as the mean and standard deviation (SD) of two or three independent experiments and the t-test was used for statistical analysis of data changes. P values of 1 were considered statistically significantly less than 0.05. data were analyzed using SPSS 18.0 and PRISM.
RESULTS
Breast tumor cells for transfection and isolation of engineered exosomes
After breast tumor cells were cultured, the cells were transfected with the miR-144 precursor sequence as described. The supernatant of transfected cells and the control cell group was collected at different passages and used for exosome isolation. The morphology of the MDA-MB-231 breast tumor cell lines by contrast phase microscopy after transfection of the miRNA precursor sequence is shown in Figure 1.
Figure 1. Morphology of MDA-MB-231 Breast Tumor Cells by Contrast Phase microscopy.
Melting curve analysis
In this study, melting curve analysis was used to confirm the accuracy of the amplified fragment and to ensure the absence of nonspecific product, primer-dimer, and contamination. By examining the generated peaks, it can be concluded that the peaks formed at low temperatures are directly related to the amount of nonspecific products formed at the end of the PCR process. As shown in Figure 2, the presence of only one peak for each gene at its specific melting temperature indicates the specificity of the replication product. This ensures no such thing as nonspecific replication, primer dimers, or contamination.

Figure 2. Characterization (specificity) amplification in Real-time PCR reaction. Each peak for the mir-144, smad4, TGF-bR2 and GAPDH genes represents the melting temperature of a PCR product.
Regulation pattern of miR-144
Following the transfection of breast cancer cells with miR-144 precursor sequence, Real-time PCR results in two transfected, and untransfected cell lines (control group) showed that miR-144 expression in transfected cells was compared. There was a significant increase in the control group. As shown below, after 24h of transfection with the precursor sequence, miR-144 expression levels were significantly increased in transfected cells compared to basal miRNA expression levels in the control cell group (p <0.001) evidence of the adequacy of the strategy chosen to increase miR-144 expression. Figure 3 shows the expression of miR-144 from real-time PCR.

Figure 3. Expression of miR-144 from Real-time PCR. Significant increase in miR-144 expression levels in cells transfected with the miR-144 precursor sequence compared to its basal expression levels in the control cell group.
Size and Morphology of Exosomes Isolated from Breast Cancer Cells by Scanning Electron Microscopy
SEM assessed the size and morphology of exosomal extracellular vesicles. The results showed that the isolated exosomes had less than 150 nm spherical appearance. Figure 4 shows the image taken by this microscope.

Figure 4. Investigation of exosomal extracellular vesicles isolated by SEM.
Evaluation of relative amounts of miR-144 in engineered exosomes compared to control exosomes
Real-time PCR was used to demonstrate the production of engineered exosomes containing miR-144. As shown, engineered exosomes (Exo-miR-144) showed significantly higher expression of miR-144 than control exosomes (p <0.001). Figure 5 shows the expression levels of miR-144 in the two engineered and control exosomes.
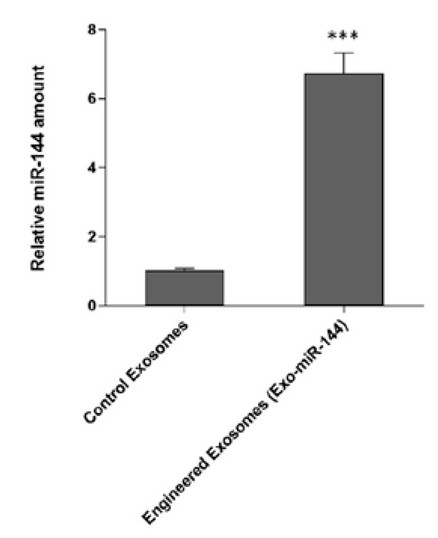
Figure 5. Determination of expression levels of miR-144 in the two engineered exosomes and control showed significant levels of miR-144 in the exo-miR-144 engineered exosomes. Exosomes are derived from cells transfected with the miR-144 precursor sequence.
Assessment of SMAD4 gene transcript expression in breast cancer cells following treatment with the bacterial toxin, engineered exo-miR-144 exosomes, and combination treatment of toxin A and exo-miR-144
To identify the effect of engineered exosomes containing miR-144 (Exo-miR-144) on breast cancer cells, the cells were treated with the engineered exosomes derived from both transfected and Non-transfected breast cancer cells (compared). Real-time PCR results as shown in figure 6 a decrease in Exo-miR-144 at 24 and 48 hours after treatment with SMAD4 gene expression. The results also showed that bacterial toxin A-induced reduced the expression of the SMAD4 gene in breast cancer cells. More importantly, the combination treatment of bacterial toxin A and Exo-miR-144 was engineered to create a time-dependent synergism to reduce SMAD4 gene expression.

Figure 6. Assessment of SMAD4 Transcript Expression in Breast Cancer Cells Following Treatment with Exosomal Control, Bacterial Toxin A, Exo-miR-144 Engineered Exosomes and Toxin A + Combined Treatment Exo-miR-144. Treatment of engineered exosomes containing miR-144 resulted in a significant decrease in the expression level of the SMAD4 gene. In this regard, the synergistic effect of the bacterial toxin A + Exo-miR-144 significantly reduced the expression of SMAD4 transcripts in a time-dependent manner. The results were normalized to GAPDH reference gene expression.
Assessment of TGF-βR2 gene transcript expression in breast cancer cells following treatment with bacterial toxin A engineered exo-miR-144 exosomes and combination treatment of toxin A and exo-miR-144
To identify the effect of engineered exosomes containing miR-144 (Exo-miR-144) on breast cancer cells, these cells were treated with these engineered exosomes and with cells treated with the control exosomal group), exosomes derived Non-transfected breast cancer cells (compared). Real-time PCR results, as shown in Fig. 7 showed that at 24 and 48 h after TGF-βR2 gene expression was decreased in the Exo-miR-144 treated group. The results also showed that bacterial toxin A-induced a decrease in the expression of the SMAD4 gene in breast cancer cells. But more importantly, the combination treatment of bacterial toxin A and Exo-miR-144 was engineered to produce a time-dependent synergism to reduce TGF-βR2 gene expression.
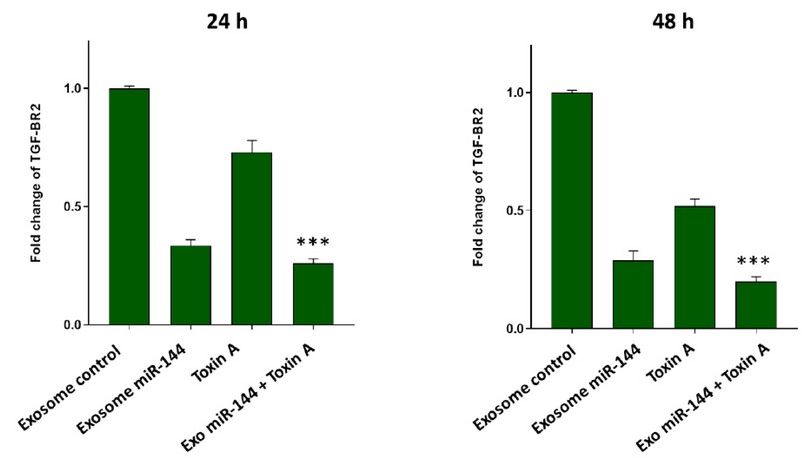
Figure 7. Evaluation of TGF-βR2 Transcript Expression in Breast Cancer Cells Following Treatment with Exosomal Control, Bacterial Toxin A, Exo-miR-144 Engineered Exosomes and Toxin A + Combined Treatment Exo-miR-144. Treatment of engineered exosomes containing miR-144 resulted in a significant decrease in the expression level of the TGF-βR2 gene. The synergistic effect of Exo-miR-144 + bacterial A toxin was important in this regard, which significantly reduced the expression of TGF-βR2 transcripts in a time-dependent manner. The results were normalized to GAPDH reference gene expression.
Evaluation of cell cycle progression in breast cancer cells following combination treatment of bacterial toxin and engineered exosomes (Exo-miR-144)
A strong synergy was observed in the reduction of expression of SMAD4 and TGF-βR2 genes following the combination treatment of bacterial toxin A and engineered exosomes (Exo-miR-144) in MDA-MB-231 breast cancer cells. As can be seen, the simultaneous treatment of bacterial toxin A and engineered exosomes (Exo-miR-144) increased the cell population in the Sub-G1 phase from 1.92% to 35.54%. This combination treatment is remarkable for stopping the cell cycle (Figure 8).
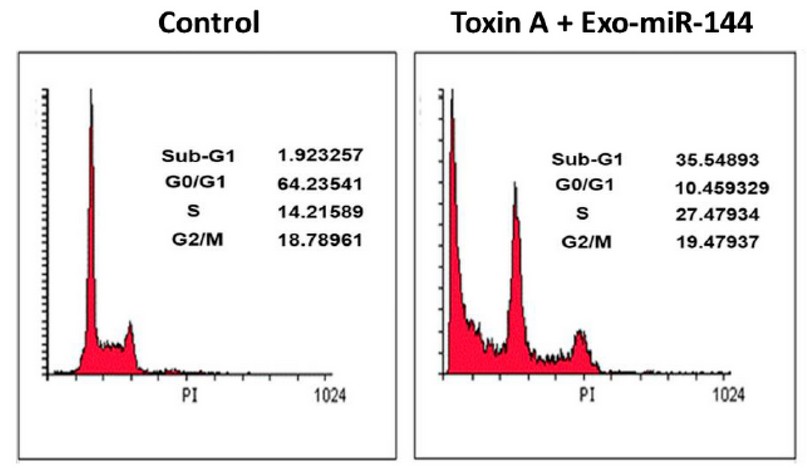
Figure 8. Cell cycle distribution in the breast cancer cell line treated with bacterial toxin A and engineered exosomes (Exo-miR-144). As shown, the suppression of TGF-β pathway gene expression increased the amount of subG1 phase cells.
DISCUSSION
Some bacterial toxins' role in inhibiting cancer cell proliferation has been elucidated. The most well-known function of Botulinum toxin is its effects on cell integrity and cytoskeleton. In 2013, Bandala and colleagues demonstrated the effect of Botulinum toxin on the proliferation and apoptosis of T47D cancer cells. They determined that Botulinum treatment may be a standard treatment for breast cancer. However, the molecular pathways induced by Botulinum that affect its cytotoxic activity have not been well identified6.
Delivery of efficient and functional miRNA mimics and/or antagonists to tumor cells is a major challenge in miRNA-based cancer therapy. The current standard approach for gene-based and RNAi-based treatments uses viral or non-viral vector systems. Methods that directly utilize viral vectors lead to efficient gene transfer, although in some cases, they also have inefficiencies7. Cationic liposomes are composed of two positively charged lipid layers. They can form with negative DNA, which has a negative charge, through the simple mixing of lipid and complex DNA so that the resulting complex (lipoplex) has a generally positive control. The lipoplex is readily attached to the cell and transfected efficiently to a large extent8. Currently, numerous clinical trials are underway using cationic liposomes to deliver genes. Liposomes for delivery of chemotherapeutic agents such as Doxorubicin have also been previously available for chemotherapy of breast cancer9. An essential drawback of using cationic liposomes is that they lack specificity for the tumor and have relatively low transfection efficiency compared to viral vectors. However, the tumor specificity of lipoplexes can be dramatically enhanced by carrying a ligand identified by the cell surface receptor. Endocytosis mediated by the cellular entry pathway receptor is highly efficient in eukaryotic cells. Ligand placement on the lipoplex facilitates the entry of DNA into cells by the initial binding of the ligand to the cell surface receptor and subsequent entry of the lipoplex into the cell. Upon entry into the cell, DNA exits the endocytosis pathway to express it in the cell nucleus10. To efficiently deliver DNA to the cell, tumor-specific nanoparticle-specific lipopolysaccharides have been developed with ligand-targeting and self-assembling capabilities for cancer gene therapy11. Exosomes are one of these biological nanostructures that efficiently transfer macromolecules and nucleic acids between cells. These natural nanofluids in the human body transfer proteins, types of RNAs, and in some cases, DNA from DNA. Cell by cell is responsible12.
In 2013, Katakowski et al. used exosomes secreted by mesenchymal stem cells to load miR-146b, showing that miRNA treatment using exosomes can effectively inhibit tumor growth. In addition to siRNAs and miRNAs, mRNAs can also be transported by exosomes as a product13. Exosomes can also load other types of medicine. For example, in 2015, Zhang et al. found that curcumin, loaded in murine lymph node cell exosomes, could successfully transfer to brain tissue and improve the apoptosis of microglial cells in the brain. The results of their work show that this strategy may provide a new non-invasive and therapeutic approach to treating inflammatory brain diseases. There are also other studies on the effect of the genetic content of exosomes in cancer treatment, all of which confirm the efficacy of exosomes in gene transfer14.
CONCLUSIONS
As shown in the results section, exosomes secreted from tumor cells did not affect the expression levels of TGF-B pathway genes in breast cancer cells. However, when miR-144 was overexpressed in tumor cells, this overexpression caused miR-144 to be packaged in tumor-derived exosomes. Interestingly, exosomes derived from these engineered cells that contained significant amounts of miR-144 reduced the expression of the SMAD4 and TGF-BR2 genes. Also, bacterial toxin A's inhibitory effects on these genes' expression were observed in a time-dependent manner. What is essential is that the exosomal combination of miR-144 and Botulinum toxin A has a synergistic effect on the expression of the SMAD4 and TGF-B genes, which may underline the importance of this therapeutic strategy.
Conflict of interest: The authors declare no conflict of interest.
REFERENCES
1. Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nature reviews cancer. 2009;9(6):400-14.
2. Cardoso F, Loibl S, Pagani O, Graziottin A, Panizza P, Martincich L, et al. The European Society of Breast Cancer Specialists recommendations for the management of young women with breast cancer. Eur J Cancer. 2012;48(18):3355-77. DOI: 10.1016/j.ejca.2012.10.004
3. Kudela E, Samec M, Kubatka P, Nachajova M, Laucekova Z, Liskova A, et al. Breast Cancer in Young Women: Status Quo and Advanced Disease Management by a Predictive, Preventive, and Personalized Approach. Cancers (Basel). 2019;11(11). doi: 10.3390/cancers11111791
4. Pollan M. Epidemiology of breast cancer in young women. Breast Cancer Res Treat. 2010;123 Suppl 1:3-6. DOI: 10.1007/s10549-010-1098-2
5. Turnbull C, Rahman N. Genetic predisposition to breast cancer: past, present, and future. Annu Rev Genomics Hum Genet. 2008;9:321-45. DOI: 10.1146/annurev.genom.9.081307.164339
6. Bakhuizen JJ, Velthuizen ME, Stehouwer S, Bleiker EM, Ausems MG. Genetic counselling of young women with breast cancer for Li-Fraumeni syndrome: a nationwide survey on the experiences and attitudes of genetics professionals. Fam Cancer. 2019;18(2):231-9. doi: 10.1007/s10689-018-0103-5.
7. Gomez-Flores-Ramos L, Castro-Sanchez A, Pena-Curiel O, Mohar-Betancourt A. Molecular Biology In Young Women With Breast Cancer: From Tumor Gene Expression To DNA Mutations. Rev Invest Clin. 2017;69(4):181-92. doi: 10.24875/ric.17002225.
8. Hua HB, Yan TT, Sun QM. miRNA polymorphisms and risk of gastric cancer in Asian population. World J Gastroenterol. 2014;20(19):5700-7. doi: 10.3748/wjg.v20.i19.5700
9. Baranwal S, Alahari SK. miRNA control of tumor cell invasion and metastasis. Int J Cancer. 2010;126(6):1283-90. doi: 10.1002/ijc.25014
10. Wang YW, Shi DB, Chen X, Gao C, Gao P. Clinicopathological significance of microRNA-214 in gastric cancer and its effect on cell biological behaviour. PLoS One. 2014;9(3):e91307. doi: 10.1371/journal.pone.0091307
11. Tsai HP, Huang SF, Li CF, Chien HT, Chen SC. Differential microRNA expression in breast cancer with different onset age. PLoS One. 2018;13(1):e0191195. doi: 10.1371/journal.pone.0191195.
12. Konoshenko MY, Lekchnov EA, Vlassov AV, Laktionov PP. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. Biomed Res Int. 2018;2018:8545347. doi: 10.1155/2018/8545347.
13. Pullmann M, Hergarten S, Laube N. Influence of a variable differential function on the stone-growth-related urinary depletion effect. Clin Chem. 2004;50(9):1675-8. doi: 10.7860/JCDR/2017/29465.10561.
14. Bandala C, Perez-Santos JL, Lara-Padilla E, Delgado Lopez G, Anaya-Ruiz M. Effect of botulinum toxin A on proliferation and apoptosis in the T47D breast cancer cell line. Asian Pac J Cancer Prev. 2013;14(2):891-4. doi: 10.7314/apjcp.2013.14.2.891.
Received: May 2 , 2022 / Accepted: June 12, 2022 / Published:15 November 2022
Citation: Simin P T, Marandi S J, Ardakani R B . MiR-144 as a novel biomarker in breast cancer diagnosis and treatment. Revis Bionatura 2022;7(4) 58. http://dx.doi.org/10.21931/RB/2022.07.04.58
